Part 3. Compare simulations
from pathlib import Path
%matplotlib inline
import matplotlib.pyplot as plt
plt.rc("figure", dpi=80)
def get_values(sim):
N = sim.params.N
Uc = sim.params.forcing.milestone.movement.periodic_uniform.speed
Dc = sim.params.forcing.milestone.objects.diameter
Lf = sim.params.forcing.milestone.movement.periodic_uniform.length
Fhc = Uc / (N * Dc)
Rec = Uc * Dc / sim.params.nu_2
period = sim.forcing.get_info()["period"]
t_statio = period
averages = sim.output.spatial_means.get_dimless_numbers_averaged(
tmin=t_statio
)
U2 = averages["dimensional"]["Uh2"]
epsK = averages["dimensional"]["epsK"]
Gamma = averages["Gamma"]
Fh = averages["Fh"]
R2 = averages["R2"]
R4 = averages["R4"]
return N, Uc, Dc, Lf, Fhc, Rec, U2, epsK, Gamma, Fh, R2, R4
import fluidsim as fls
path_dir_data = Path(fls.FLUIDSIM_PATH) / "tutorial_parametric_study"
path_runs = sorted(path_dir_data.glob("*"), key=lambda p: p.name)
[p.name for p in path_runs]
['ns3d.strat_144x144x48_V4.5x4.5x1.5_N0.2_Lf3.5_U0.01_D0.5_2021-10-05_15-35-47',
'ns3d.strat_144x144x48_V4.5x4.5x1.5_N0.2_Lf3.5_U0.02_D0.5_2021-10-05_16-12-36',
'ns3d.strat_144x144x48_V4.5x4.5x1.5_N0.2_Lf3.5_U0.04_D0.5_2021-10-05_16-49-31',
'ns3d.strat_144x144x48_V4.5x4.5x1.5_N0.2_Lf3.5_U0.06_D0.5_2021-10-05_17-25-48',
'ns3d.strat_144x144x48_V4.5x4.5x1.5_N0.2_Lf3.5_U0.08_D0.5_2021-10-05_18-01-23']
Compare simulations with Pandas
import numpy as np
from pandas import DataFrame
# fmt: off
columns = ["N", "Uc", "Dc", "Lf", "Fhc", "Rec", "U2", "epsK", "Gamma", "Fh", "R2", "R4"]
# fmt: on
values = []
for path in path_runs:
sim = fls.load(path, hide_stdout=True)
values.append(get_values(sim))
df = DataFrame(values, columns=columns)
df["min_R"] = np.array([df.R2, df.R4]).min(axis=0)
df
| N | Uc | Dc | Lf | Fhc | Rec | U2 | epsK | Gamma | Fh | R2 | R4 | min_R | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.2 | 0.01 | 0.5 | 3.5 | 0.1 | 5000.0 | 0.000030 | 5.564985e-08 | 0.300576 | 0.009451 | 1.391246 | 0.170957 | 0.170957 |
| 1 | 0.2 | 0.02 | 0.5 | 3.5 | 0.2 | 10000.0 | 0.000109 | 3.917927e-07 | 0.468731 | 0.018019 | 9.794817 | 2.204561 | 2.204561 |
| 2 | 0.2 | 0.04 | 0.5 | 3.5 | 0.4 | 20000.0 | 0.000419 | 2.834370e-06 | 0.618684 | 0.034420 | 70.859251 | 30.298269 | 30.298269 |
| 3 | 0.2 | 0.06 | 0.5 | 3.5 | 0.6 | 30000.0 | 0.000914 | 8.980284e-06 | 0.629455 | 0.049961 | 224.507095 | 139.360780 | 139.360780 |
| 4 | 0.2 | 0.08 | 0.5 | 3.5 | 0.8 | 40000.0 | 0.001652 | 2.199885e-05 | 0.561550 | 0.067149 | 549.971217 | 463.932956 | 463.932956 |
df["log10_R2"] = np.log10(df.R2)
fig, ax = plt.subplots()
df.plot.scatter(ax=ax, x="Fh", y="Gamma", c="log10_R2", cmap="plasma");
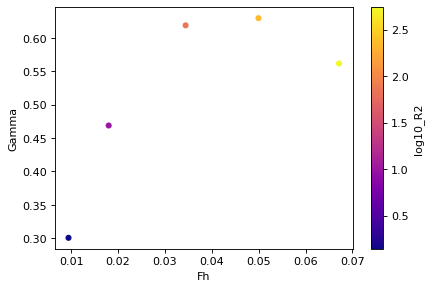
A figure with data from different simulations
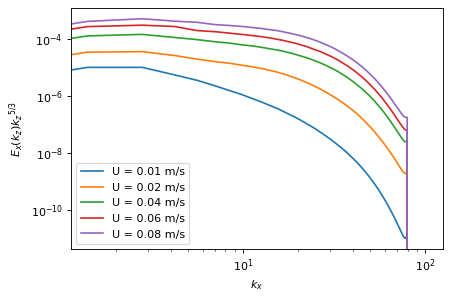
fig, ax = plt.subplots()
for path in path_runs:
sim = fls.load(path, hide_stdout=True)
period = sim.forcing.get_info()["period"]
data = sim.output.spectra.load1d_mean(tmin=period)
spectrum = data["spectra_vx_kx"]
kx = data["kx"]
ax.loglog(
kx,
spectrum * kx ** (5 / 3),
label=f"U = {sim.params.forcing.milestone.movement.periodic_uniform.speed} m/s",
)
ax.set_xlabel("$k_x$")
ax.set_ylabel("$E_x(k_z) {k_z}^{5/3}$")
plt.legend();
compute mean of spectra
tmin = 800.502 ; tmax = 1600.67
imin = 100 ; imax = 200
compute mean of spectra
tmin = 400.051 ; tmax = 800.132
imin = 100 ; imax = 200
compute mean of spectra
tmin = 200.093 ; tmax = 400.025
imin = 100 ; imax = 200
compute mean of spectra
tmin = 133.342 ; tmax = 266.703
imin = 101 ; imax = 201
compute mean of spectra
tmin = 100.004 ; tmax = 200.024
imin = 100 ; imax = 200